Spring 2021 eNewsletter
Spotlight: Environmental DNA and RNA for advanced management and accurate surveillance of aquatic invasive species in the Great Lakes
The Cooperative Institute for Great Lakes Research (CIGLR) in partnership with NOAA-GLERL has been working to develop genomic tools that will advance conservation biology and biodiversity management. Drs. Nathaniel Marshall (CIGLR), Henry Vanderploeg (GLERL), and Rao Chaganti (CIGLR) recently published an article in Scientific Reports describing a new methodology they developed that uses environmental DNA and RNA (eDNA and eRNA) to accurately detect and distinguish the presence of living organisms and improve estimates of their relative abundances. eDNA and eRNA refer to genetic material naturally released from an organism into the environment, through feces/urine, mucus, gametes, skin/tissue, carcasses, and so on. It can be collected from the environment (such as lake water), rather than directly from an organism, and analyzed to identify the species present.
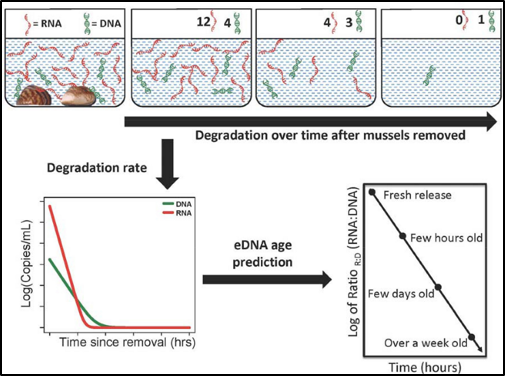
The concentration of eRNA depletes faster than eDNA, providing a predictor for estimating time since genomic material release.
For successful management of bio-invasions and monitoring the reintroduction of threatened or endangered species in the Great Lakes or elsewhere, managers and stakeholders need timely and robust scientific advice. Traditionally, scientists have relied on conventional approaches such as visual surveys to measure species presence, but low rates of detection can increase the cost and labor required for this approach. During the last decade, eDNA has become a powerful tool used to collect information from aquatic habitats. eDNA methodology is cost-effective and easier to collect compared to traditional sampling, and has improved the management and assessment of species distribution. eDNA analysis has been incorporated into Great Lakes monitoring programs for detection of a wide range of organisms, including species that are endangered/threatened, invasive, or economically important.
Although eDNA is a rapid and robust methodology, some concerns still exist in current eDNA protocols. False positives (i.e., detection of species not actually present) are sometimes a concern with eDNA results. False positives occur when eDNA is naturally transported from one environment to another, such as with currents, natural dispersion within the water column, or from predator and/or human mediated movement. False positives can obscure species’ spatial patterns and prevent true understanding of community assemblages. These drawbacks often raise major concerns for stakeholders and environmental managers working toward resource mitigation.
To address these concerns, Marshall, Vanderploeg, and Chaganti developed a new methodology that uses eDNA and eRNA to accurately detect species presence and estimate relative abundance. Because the concentration of eRNA degrades much faster than the eDNA, it provides a predictor for estimating time since genomic material was released from an organism into the environment, thus improving spatiotemporal estimates of community composition. This new method is easily modified and can be applied to monitoring different species within the Great Lakes or elsewhere in the world. It is a powerful technique that will enhance the confidence of rapid decision-making by environmental managers and stakeholders and expand monitoring of ecosystem health.
Read the recently published article:
Marshall, Nathaniel T., Henry A. Vanderploeg & Subba Rao Chaganti. 2021. Environmental (e)RNA advances the reliability of eDNA by predicting its age. Scientific Reports. 11:2769. (DOI:10.1038/s41598-021-82205-4).
