Fall 2019 eNewsletter
Featured Research
Using Environmental DNA and RNA to Inform Biomonitoring and Bioassessment Management Plans

Dr. Nate Marshall sampling eDNA and eRNA from a mesocosm experiment. Photo Credit: Aubrey Lashaway.
Identifying species distributions and understanding changes in community composition are crucial for mitigating the adverse effects of invasive species. The Great Lakes ecosystem is currently home to 188 aquatic non-native species, 55 of which are invasive and cause ecological and economic harm. With growing concerns over potential new invaders (e.g., silver and bighead carp), early detection and continued monitoring of aquatic invasive species remain a high priority for management agencies in the United States.
Nate Marshall is a CIGLR postdoctoral research fellow working with Drs. Hank Vanderploeg (NOAA GLERL), Ed Rutherford (NOAA GLERL), and Rao Chaganti (CIGLR) to develop new techniques to improve ecological understanding and environmental monitoring of invasive species, foodweb dynamics, and plankton community composition in the Great Lakes.
“I am developing environmental DNA/RNA (eDNA/eRNA) techniques to inform biomonitoring and bioassessment management plans,” says Nate.
eDNA refers to is genetic material naturally released from an organism into the environment, through feces/urine, mucus, gametes, skin/tissue, carcasses, and so on. It can be collected from the environment (such as lake water), rather than directly from an organism, and analyzed to identify the species present.
“This technique detects an organism’s presence without it ever being physically seen,” says Nate. “Using eDNA methods can improve early detection of invasive species, which tend to go unrecognized until their populations become an ecological or economic problem.”
eDNA analysis is a sensitive method for revealing rare species, and has been incorporated into Great Lakes monitoring programs for the detection of Asian carp. However, some experimental flaws still exist in current eDNA protocols.

Nate is using invasive zebra and quagga mussels in mesocosm experiments (pictured above) to explore how much eDNA and eRNA is shed into their surrounding environment and how long it persists. Photo Credit: Nate Marshall.
“False positives (i.e., detection of species not actually present) are sometimes a concern with eDNA results, which could result in misuse of mitigation resources,” says Nate. “False positives occur when eDNA is naturally transported from one environment to another, such as with currents, natural dispersion within the water column, or from predator and/or human mediated movement. False positives can obscure species spatial patterns and prevent true understanding of community assemblages.”
Recently, eDNA methods have been used for assessing the status of invasive zebra and quagga mussels for many water bodies around the Great Lakes and in western U.S. states. Quantifying eDNA alone may not fully represent the actual species abundance in an environment. However, eRNA degrades faster and including this method may reduce false positives and thus improve spatiotemporal estimates of community composition.
Currently, Nate and his team are using mesocosm experiments to explore how much eDNA and eRNA invasive dreissenids (zebra and quagga mussels) shed into their environment and how long it persists.
“By sampling a combination of eDNA and eRNA, we hope to better elucidate the presence of living organisms within a location and improve estimators of their relative abundances. Our mesocosm experiments will provide evidence for collection and quantification of eRNA shed from organisms, rather than just analyzing for eDNA,” says Nate.
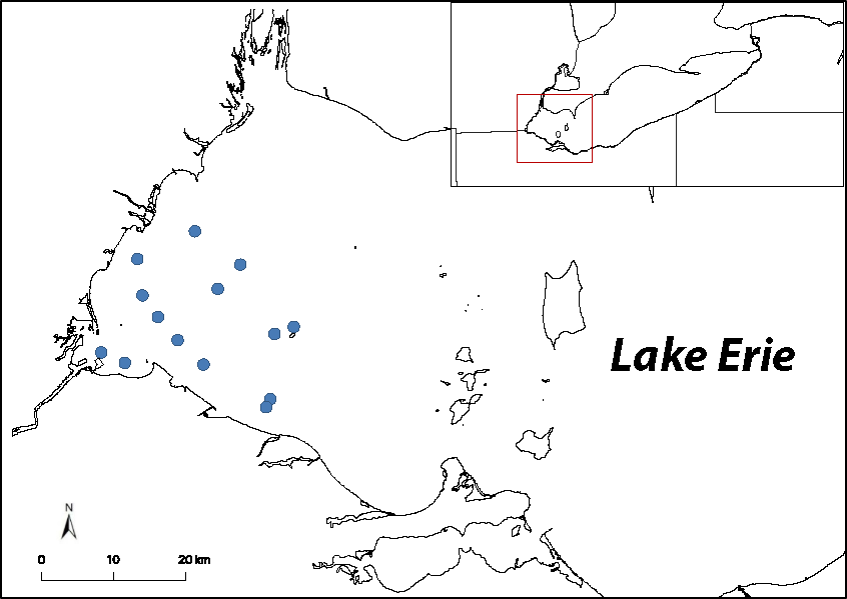
This past summer (2019), water samples were collected from 14 stations (blue dots) in the western basin of Lake Erie, and are being analyzed for eDNA and eRNA to determine if either molecule accurately displays the known distribution and abundance patterns of dreissenid mussel beds. Image Credit: Nate Marshall.
Preliminary results from these experiments display faster degradation of RNA molecules, indicating RNA is a better gauge of an organism’s recent activity within the environment. This past summer (2019), water samples were collected from 14 stations in the western basin of Lake Erie, where dreissenid mussel beds vary in their abundance. Currently, these water samples are being analyzed for eDNA and eRNA to determine if either molecule accurately displays the known distribution and abundance patterns of these mussel beds.
“I believe eRNA will address fundamental and applied research questions within ecology and conservation by exploring bioassessment of aquatic ecosystems through ecogenomic approaches. Our goal is to establish methods for utilizing eRNA in future biomonitoring programs that would concurrently detect living organisms and provide information on the stress-level of those organisms. I hope this can develop into a powerful technique for estimating ecosystem health,” says Nate.
About the Author
Previously, Nate used a combination of field studies and mesocosms to explore the utility of eDNA for describing invasive species populations and macroinvertebrate community composition. He received his B.S. from the Ohio State University (2010) in Zoology, M.S. from the University of Texas at Tyler (2014) in Biology, and Ph.D. from the University of Toledo (2019) in Environmental Sciences.
